The Xie laboratory uses Drosophila and mice to study stem cell-niche interactions, self-renewal, differentiation, quality control, and aging. Here are some research accomplishments and future directions for stem cell research in the Xie laboratory.
1. Niche and intrinsic control of stem cell self-renewal
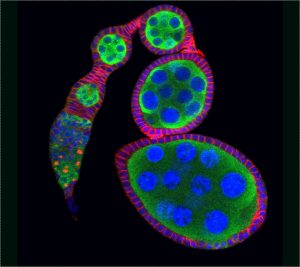
A stem cell can divide both asymmetrically and symmetrically. Regarding the former, one renewing stem cell and one differentiating daughter cell is generated, whereas with regards to the latter either two renewing stem cells or two differentiating daughters are produced. To investigate how stem cell fate is determined, Dr Xie was the first to use germline stem cells (GSCs) in the Drosophila ovary and demonstrate that stem cell self-renewal is controlled by the niche (Science, 2000). Since then, the Xie laboratory has shown that cap cells and GSC-contacting inner germarial sheath (IGS) cells contribute to the niche for controlling self-renewal (Science, 2000; Development, 2007; PLoS Genet, 2011; Curr Biol, 2021). His laboratory has also demonstrated that: 1) Dpp/BMP is a key niche signal, which functions at a short range to control GSC self-renewal by directly repressing differentiation (Cell, 1998; Development, 2003). 2) E-cadherin anchors stem cells in the niche, and it is also directly linked to stem cell competition and quality control (Science, 2002; PNAS, 2002; Cell Stem Cell, 2008). 3) Notch signaling directly controls niche formation and size (Development, 2007). 4) stem cell aging is controlled by various niche signals, including BMP (Cell Stem Cell, 2007). These findings have provided novel insights into how the niche controls stem cell self-renewal, competition, and aging.
For niche signals to control stem cell self-renewal, it was hypothesized that they must directly regulate the expression and functions of various intrinsic factors in stem cells. The Xie laboratory has identified various classes of intrinsic factors that control GSC self-renewal by distinct mechanisms. These include: 1) microtubule-binding protein Lissencephaly-1 (Lis-1) and epigenetic factor ISWI, which promote BMP signaling and E-cadherin-mediated cell adhesion (PNAS, 2010; Science, 2005). 2) Translation initiation factor eIF4A and CSN4, which interact with Bam to inactivate its function and thus repress differentiation (PNAS, 2009; Nature, 2014). 3) miRNA component Dcr-1, translation regulator Pelota, H3K9 trimethylase Eggless and 5mC RNA binding protein Yps, which repress Bam-independent differentiation pathways (Development, 2005; Curr Biol, 2007; PLoS Genet, 2011; PNAS, 2020). In addition, DNA damage was shown to disrupt GSC self-renewal primarily through Chk2 activation (Cell Reports, 2015; Development, 2015; Developmental Cell, 2017). These findings provide new insight into how intrinsic factors work with the niche to control GSC self-renewal. We are currently expanding our studies to include spermatogonial stem cells and their niches in the mouse testis.
Research directions:
- To identify niche components for controlling GSC self-renewal in the mouse testis.
- To investigate how niche-derived factors control GSC self-renewal by interacting with intrinsic factors in Drosophila and mice.
- To investigate how various RNA-binding proteins work cooperatively to control GSC self-renewal intrinsically in Drosophila and mice.
- To investigate how GSC aging is controlled extrinsically and intrinsically in Drosophila and mice.
2. Niche and intrinsic control of stem cell lineage differentiation
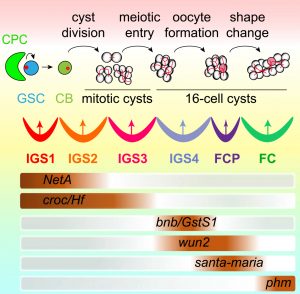
Contrary to the conventional thinking that stem cell progeny differentiation is a developmental default state, the Xie laboratory demonstrated that GSC progeny differentiation is also controlled by the niche, which they named the differentiation niche (Development, 2011; PLoS Genetics, 2013). This niche is formed by posterior IGS cells and is regulated by the Hh, Wnt, and EGFR signaling pathways to promote GSC progeny differentiation at least in part by repressing BMP signaling (Development, 2011, 2015; eLife, 2015; Science Advances, 2020). Furthermore, they showed that the COP9 signalosome, exocyst complex, Rho1, and H3K9 trimethylase Eggless are all required in the niche to regulate Hh, Wnt, EGFR, or BMP signaling for controlling differentiation of the GSC progeny (Development, 2011, 2019; PLoS Genet, 2011). In addition, more recently this niche was shown to contain multiple compartments for controlling germ cell cyst formation, meiotic entry, and oocyte specification/germline cyst shape change (Curr Biol, 2021). Therefore, the differentiation of GSC progeny is programmed step-by-step by the niche. We now aim to extend our findings on the differentiation niche in Drosophila, and investigate mammalian systems.
As stem cell self-renewal is controlled intrinsically, the Xie laboratory has also demonstrated that intrinsic factors work within GSC progeny. These stimulate differentiation by repressing the expression or functions of self-renewal factors, which promote the expression or functions of other differentiation factors. They showed that: 1) master differentiation factor Bam interacts with Bgcn and eIF4A to form a ternary protein complex to repress the translation of self-renewal factors and promote the expression of differentiation factors (PNAS, 2009). 2) Bam can also convert the self-renewal-promoting functions of the COP9 and CCR4-Not complexes into the differentiation-promoting subcomplexes via protein competition (Nature, 2014; Cell Reports, 2015). 3) piRNA component Aubergine and 5mC RNA binding protein Yps promote the expression of self-renewal factors and repress the expression of differentiation factors (Developmental Cell, 2017; PNAS, 2020). These findings have provided novel insights into how intrinsic factors control GSC progeny differentiation.
Research directions:
- To define the niche for controlling GSC progeny differentiation in the mouse testis.
- To investigate how niche compartments control stepwise GSC progeny differentiation in Drosophila and mice.
- To investigate how niche signals work with intrinsic factors to control GSC progeny differentiation in Drosophila and mice.